ViTAE:引入归纳偏置,浅层用CNN,深层用self-attention
共 18817字,需浏览 38分钟
·
2022-02-13 14:09
【GaintPanda导语】
引入归纳偏置,即局部性和尺度不变性,浅层用CNN编码token,深层用多头注意力机制做transformer的模块堆叠,是一次CNN和transformer结合探索,也是一个对送进多头注意力机制的token编码的探索,送进更信息更加聚合的token,来降低模型大小,提高分类任务的效果。
引言
一句话总结:浅层用CNN,深层用self-attention,文字写得非常好,通俗易懂,亮点一般。
主观讨论,ViTAE实际上是做了token编码,这点跟T2T的思路是一样。引入CNN的原因:引入两种归纳偏置,即局部性和尺度不变性。
其实已经有很多transformer的工作指出了,引入CNN能够提升transformer的性能和训练稳定性;这点严格来说并不能算做novelty,因为是同期工作,不排除撞车的情况。
论文名词:《ViTAE: Vision Transformer Advanced by Exploring Intrinsic Inductive Bias》
出处:NeurIPS 2021 陶大程团队
Locality局部性: CNN通过计算局部相邻的像素点之间的联系能够提取局部特征,比如边缘和角,能在浅层提供丰富局部特征(local feature),而这些浅层信息能进一步聚合提取深层语义信息在接下来的一系列的CNN。
Scale-invariant尺度不变性:CNN能通过分层结构提取多尺度特征(multi-scale features),因此尺度不变性能通过空洞卷积和层间或层内的特征融合 (intra- or inter-layer feature fusion)体现出来。
网络结构:
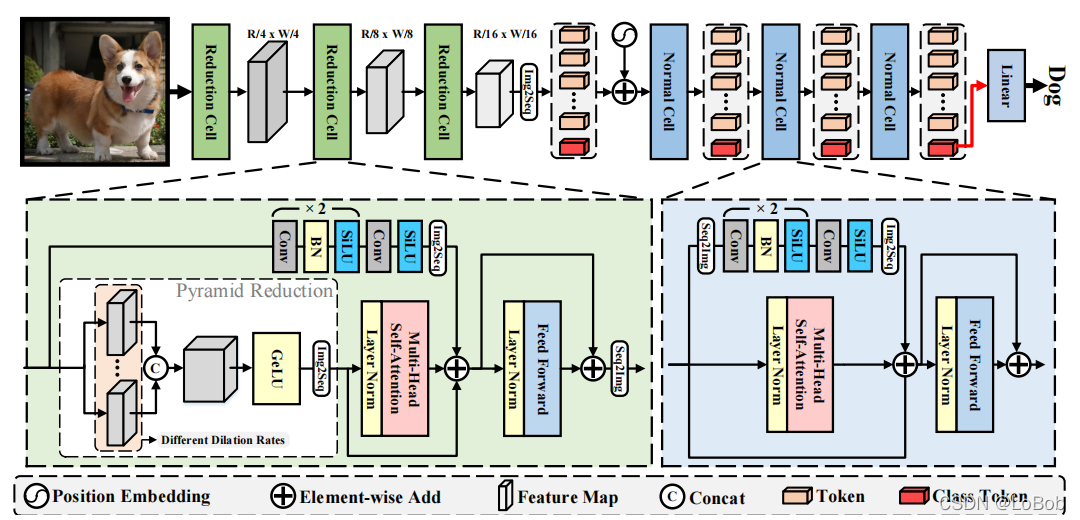
1、ViTAE采用分治的思想,分别建模局部关系和长距离依赖关系,再将其融合起来。
2、两个模块:Reduction Cell (RC) and Normal Cell (NC)。Reduction Cell做浅层编码和降采样生成token,Normal Cell联合建模token中的局部性和全局依赖关系。
3、RC和NC的区别:img2seq,NC没有Pyramid Reduction Module(PRM) 。
4、Pyramid Reduction Module(PRM):就是用空洞卷积实现,然后concat起来。这里是为了做尺度不变性。
5、接着将concat的结果做多头注意力Multi-head self-attention。
6、在对没做PRM的feature map做一系列卷积操作,称为Parallel Convolutional Module (PCM) ,这里是为了提取局部特征。NC模块中,class token在PCM模块计算中被丢弃,因为class token跟其他token没有空间信息联系。
7、最后参考残差将他们加起来送进FFN。
8、跟ViT一样使用了class token,并加上了sinusoid position encoding(PE),这里个人觉得可以把class token去掉的,估计作者参考的工作是T2T,就沿用class token和PE。
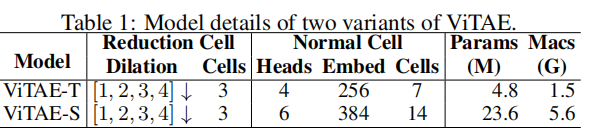
整个网络的堆叠情况如下table 1:
整体网络结构图如下图:
class PRM(nn.Module):
def __init__(self, img_size=224, kernel_size=4, downsample_ratio=4, dilations=[1,6,12], in_chans=3, embed_dim=64, share_weights=False, op='cat'):
super().__init__()
self.dilations = dilations
self.embed_dim = embed_dim
self.downsample_ratio = downsample_ratio
self.op = op
self.kernel_size = kernel_size
self.stride = downsample_ratio
self.share_weights = share_weights
self.outSize = img_size // downsample_ratio
if share_weights:
self.convolution = nn.Conv2d(in_channels=in_chans, out_channels=embed_dim, kernel_size=self.kernel_size, \
stride=self.stride, padding=3*dilations[0]//2, dilation=dilations[0])
else:
self.convs = nn.ModuleList()
for dilation in self.dilations:
padding = math.ceil(((self.kernel_size-1)*dilation + 1 - self.stride) / 2)
self.convs.append(nn.Sequential(*[nn.Conv2d(in_channels=in_chans, out_channels=embed_dim, kernel_size=self.kernel_size, \
stride=self.stride, padding=padding, dilation=dilation),
nn.GELU()]))
if self.op == 'sum':
self.out_chans = embed_dim
elif op == 'cat':
self.out_chans = embed_dim * len(self.dilations)
def forward(self, x):
B, C, W, H = x.shape
if self.share_weights:
padding = math.ceil(((self.kernel_size-1)*self.dilations[0] + 1 - self.stride) / 2)
y = nn.functional.conv2d(x, weight=self.convolution.weight, bias=self.convolution.bias, \
stride=self.downsample_ratio, padding=padding, dilation=self.dilations[0]).unsqueeze(dim=-1)
for i in range(1, len(self.dilations)):
padding = math.ceil(((self.kernel_size-1)*self.dilations[i] + 1 - self.stride) / 2)
_y = nn.functional.conv2d(x, weight=self.convolution.weight, bias=self.convolution.bias, \
stride=self.downsample_ratio, padding=padding, dilation=self.dilations[i]).unsqueeze(dim=-1)
y = torch.cat((y, _y), dim=-1)
else:
y = self.convs[0](x).unsqueeze(dim=-1)
for i in range(1, len(self.dilations)):
_y = self.convs[i](x).unsqueeze(dim=-1)
y = torch.cat((y, _y), dim=-1)
B, C, W, H, N = y.shape
if self.op == 'sum':
y = y.sum(dim=-1).flatten(2).permute(0,2,1).contiguous()
elif self.op == 'cat':
y = y.permute(0,4,1,2,3).flatten(3).reshape(B, N*C, W*H).permute(0,2,1).contiguous()
else:
raise NotImplementedError('no such operation: {} for multi-levels!'.format(self.op))
return y, (W, H)
class ReductionCell(nn.Module):
def __init__(self, img_size=224, in_chans=3, embed_dims=64, token_dims=64, downsample_ratios=4, kernel_size=7,
num_heads=1, dilations=[1,2,3,4], share_weights=False, op='cat', tokens_type='performer', group=1,
drop=0., attn_drop=0., drop_path=0., mlp_ratio=1.0):
super().__init__()
self.img_size = img_size
self.op = op
self.dilations = dilations
self.num_heads = num_heads
self.embed_dims = embed_dims
self.token_dims = token_dims
self.in_chans = in_chans
self.downsample_ratios = downsample_ratios
self.kernel_size = kernel_size
self.outSize = img_size
PCMStride = []
residual = downsample_ratios // 2
for _ in range(3):
PCMStride.append((residual > 0) + 1)
residual = residual // 2
assert residual == 0
self.pool = None
if tokens_type == 'pooling':
PCMStride = [1, 1, 1]
self.pool = nn.MaxPool2d(downsample_ratios, stride=downsample_ratios, padding=0)
tokens_type = 'transformer'
self.outSize = self.outSize // downsample_ratios
downsample_ratios = 1
self.PCM = nn.Sequential(
nn.Conv2d(in_chans, embed_dims, kernel_size=(3, 3), stride=PCMStride[0], padding=(1, 1), groups=group), # the 1st convolution
nn.SiLU(inplace=True),
nn.Conv2d(embed_dims, embed_dims, kernel_size=(3, 3), stride=PCMStride[1], padding=(1, 1), groups=group), # the 1st convolution
nn.BatchNorm2d(embed_dims),
nn.SiLU(inplace=True),
nn.Conv2d(embed_dims, token_dims, kernel_size=(3, 3), stride=PCMStride[2], padding=(1, 1), groups=group), # the 1st convolution
nn.SiLU(inplace=True))
self.PRM = PRM(img_size=img_size, kernel_size=kernel_size, downsample_ratio=downsample_ratios, dilations=self.dilations,
in_chans=in_chans, embed_dim=embed_dims, share_weights=share_weights, op=op)
self.outSize = self.outSize // downsample_ratios
in_chans = self.PRM.out_chans
if tokens_type == 'performer':
assert num_heads == 1
self.attn = Token_performer(dim=in_chans, in_dim=token_dims, head_cnt=num_heads, kernel_ratio=0.5)
elif tokens_type == 'performer_less':
self.attn = None
self.PCM = None
elif tokens_type == 'transformer':
self.attn = Token_transformer(dim=in_chans, in_dim=token_dims, num_heads=num_heads, mlp_ratio=mlp_ratio, drop=drop, attn_drop=attn_drop, drop_path=drop_path)
self.num_patches = (img_size // 2) * (img_size // 2) # there are 3 sfot split, stride are 4,2,2 seperately
def forward(self, x):
if len(x.shape) < 4:
B, N, C = x.shape
n = int(np.sqrt(N))
x = x.view(B, n, n, C).contiguous()
x = x.permute(0, 3, 1, 2)
if self.pool is not None:
x = self.pool(x)
PRM_x, _ = self.PRM(x)
if self.attn is None:
return PRM_x
convX = self.PCM(x)
x = self.attn.attn(self.attn.norm1(PRM_x))
convX = convX.permute(0, 2, 3, 1).view(*x.shape).contiguous()
x = x + convX
x = x + self.attn.drop_path(self.attn.mlp(self.attn.norm2(x)))
return x
class NormalCell(nn.Module):
def __init__(self, dim, num_heads, mlp_ratio=4., qkv_bias=False, qk_scale=None, drop=0., attn_drop=0.,
drop_path=0., act_layer=nn.GELU, norm_layer=nn.LayerNorm, class_token=False, group=64, tokens_type='transformer'):
super().__init__()
self.norm1 = norm_layer(dim)
self.class_token = class_token
if tokens_type == 'transformer':
self.attn = Attention(
dim, num_heads=num_heads, qkv_bias=qkv_bias, qk_scale=qk_scale, attn_drop=attn_drop, proj_drop=drop)
elif tokens_type == 'performer':
self.attn = AttentionPerformer(
dim, num_heads=num_heads, qkv_bias=qkv_bias, qk_scale=qk_scale, attn_drop=attn_drop, proj_drop=drop)
self.drop_path = DropPath(drop_path) if drop_path > 0. else nn.Identity()
self.norm2 = norm_layer(dim)
mlp_hidden_dim = int(dim * mlp_ratio)
self.mlp = Mlp(in_features=dim, hidden_features=mlp_hidden_dim, act_layer=act_layer, drop=drop)
self.PCM = nn.Sequential(
nn.Conv2d(dim, mlp_hidden_dim, 3, 1, 1, 1, group),
nn.BatchNorm2d(mlp_hidden_dim),
nn.SiLU(inplace=True),
nn.Conv2d(mlp_hidden_dim, dim, 3, 1, 1, 1, group),
nn.BatchNorm2d(dim),
nn.SiLU(inplace=True),
nn.Conv2d(dim, dim, 3, 1, 1, 1, group),
nn.SiLU(inplace=True),
)
def forward(self, x):
b, n, c = x.shape
if self.class_token:
n = n - 1
wh = int(math.sqrt(n))
convX = self.drop_path(self.PCM(x[:, 1:, :].view(b, wh, wh, c).permute(0, 3, 1, 2).contiguous()).permute(0, 2, 3, 1).contiguous().view(b, n, c))
x = x + self.drop_path(self.attn(self.norm1(x)))
x[:, 1:] = x[:, 1:] + convX
else:
wh = int(math.sqrt(n))
convX = self.drop_path(self.PCM(x.view(b, wh, wh, c).permute(0, 3, 1, 2).contiguous()).permute(0, 2, 3, 1).contiguous().view(b, n, c))
x = x + self.drop_path(self.attn(self.norm1(x)))
x = x + convX
x = x + self.drop_path(self.mlp(self.norm2(x)))
return x
Figure 4是grad-CAM,会有一些不同,但个人私货环节,可能并不是每个样本都会之前的方法好。

效果如下图table 2,引入了3个stage的CNN,将224*224做完token送进多头注意力机制的token数量降低了,因此参数量降低是很直观的,分类任务上效果还可以。

训练的配方:输入224x224, AdamW,用cosine learning rate scheduler,初始学习率0.0001,batch size是512, 数据增强方法跟T2T一样即mixup和cutmix。针对ViT难以训练的毛病,表现了ViTAE能够在小的数据集中收敛和训练速度,这部分在个人觉得在加入了CNN后,就会有这方面的增益。
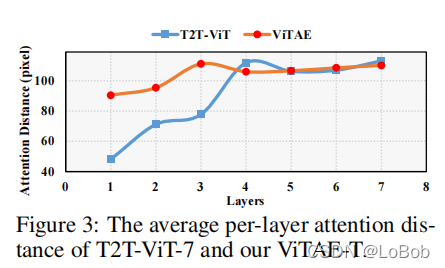
平均注意力距离:Figure 3做了一个挺有意思的分析,average attention distance of each layer,每层平均注意力距离。ViTAE在浅层(1-3层,也就是RCs)的长距离依赖关系弱于深层(即4-7层,也就是NCs)。说明了训练完的模型,确实跟一开始的设定一样。
但我不是很同意文中的表述(It can be observed that with the usage of PCM, which focuses on modeling locality, the transformer layers in the proposed NCs can better focus on modeling long-range dependencies, especially in shallow layers.),因为NC也有PCM模块。
个人思考:ConvNext将Swin Transformer一步步用CNN来替换,也获得非常不错的效果。实际上使用的时候,个人认为并不用关心它是transformer还是CNN,只要相应的算子能支持,在算力、功耗、指标等限制下,能跑得更快更好那就可以了。
ConvNext侧面证明了一点,纯transformer的架构并没有遥遥领先与CNN架构,那么如何将CNN和transformer有机的结合在一起,在更具体的任务中能够提点,是我关心的要点。
ViTAE也是CNN和tranformer结合的一种尝试,与其说是CNN结合transformer,我更愿意理解为:拿更有效的token送进self-attention中去建模。
ViTAE:引入归纳偏置,浅层用CNN,深层用self-attention
一句话总结:浅层用CNN,深层用self-attention,文字写得非常好,通俗易懂,亮点一般。
主观讨论,ViTAE实际上是做了token编码,这点跟T2T的思路是一样。引入CNN的原因:引入两种归纳偏置,即局部性和尺度不变性。
其实已经有很多transformer的工作指出了,引入CNN能够提升transformer的性能和训练稳定性;这点严格来说并不能算做novelty,因为是同期工作,不排除撞车的情况。
论文名词:《ViTAE: Vision Transformer Advanced by Exploring Intrinsic Inductive Bias》 出处:NeurIPS 2021 陶大程团队
Locality局部性: CNN通过计算局部相邻的像素点之间的联系能够提取局部特征,比如边缘和角,能在浅层提供丰富局部特征(local feature),而这些浅层信息能进一步聚合提取深层语义信息在接下来的一系列的CNN。
Scale-invariant尺度不变性:CNN能通过分层结构提取多尺度特征(multi-scale features),因此尺度不变性能通过空洞卷积和层间或层内的特征融合 (intra- or inter-layer feature fusion)体现出来。
网络结构:
1、ViTAE采用分治的思想,分别建模局部关系和长距离依赖关系,再将其融合起来。
2、两个模块:Reduction Cell (RC) and Normal Cell (NC)。Reduction Cell做浅层编码和降采样生成token,Normal Cell联合建模token中的局部性和全局依赖关系。
3、RC和NC的区别:img2seq,NC没有Pyramid Reduction Module(PRM) 。
4、Pyramid Reduction Module(PRM):就是用空洞卷积实现,然后concat起来。这里是为了做尺度不变性。
5、接着将concat的结果做多头注意力Multi-head self-attention。
6、在对没做PRM的feature map做一系列卷积操作,称为Parallel Convolutional Module (PCM) ,这里是为了提取局部特征。NC模块中,class token在PCM模块计算中被丢弃,因为class token跟其他token没有空间信息联系。
7、最后参考残差将他们加起来送进FFN。
8、跟ViT一样使用了class token,并加上了sinusoid position encoding(PE),这里个人觉得可以把class token去掉的,估计作者参考的工作是T2T,就沿用class token和PE。
整个网络的堆叠情况如下table 1:

整体网络结构图如下图:
 ViTAE 整体结构图
ViTAE 整体结构图
class PRM(nn.Module):
def __init__(self, img_size=224, kernel_size=4, downsample_ratio=4, dilations=[1,6,12], in_chans=3, embed_dim=64, share_weights=False, op='cat'):
super().__init__()
self.dilations = dilations
self.embed_dim = embed_dim
self.downsample_ratio = downsample_ratio
self.op = op
self.kernel_size = kernel_size
self.stride = downsample_ratio
self.share_weights = share_weights
self.outSize = img_size // downsample_ratio
if share_weights:
self.convolution = nn.Conv2d(in_channels=in_chans, out_channels=embed_dim, kernel_size=self.kernel_size, \
stride=self.stride, padding=3*dilations[0]//2, dilation=dilations[0])
else:
self.convs = nn.ModuleList()
for dilation in self.dilations:
padding = math.ceil(((self.kernel_size-1)*dilation + 1 - self.stride) / 2)
self.convs.append(nn.Sequential(*[nn.Conv2d(in_channels=in_chans, out_channels=embed_dim, kernel_size=self.kernel_size, \
stride=self.stride, padding=padding, dilation=dilation),
nn.GELU()]))
if self.op == 'sum':
self.out_chans = embed_dim
elif op == 'cat':
self.out_chans = embed_dim * len(self.dilations)
def forward(self, x):
B, C, W, H = x.shape
if self.share_weights:
padding = math.ceil(((self.kernel_size-1)*self.dilations[0] + 1 - self.stride) / 2)
y = nn.functional.conv2d(x, weight=self.convolution.weight, bias=self.convolution.bias, \
stride=self.downsample_ratio, padding=padding, dilation=self.dilations[0]).unsqueeze(dim=-1)
for i in range(1, len(self.dilations)):
padding = math.ceil(((self.kernel_size-1)*self.dilations[i] + 1 - self.stride) / 2)
_y = nn.functional.conv2d(x, weight=self.convolution.weight, bias=self.convolution.bias, \
stride=self.downsample_ratio, padding=padding, dilation=self.dilations[i]).unsqueeze(dim=-1)
y = torch.cat((y, _y), dim=-1)
else:
y = self.convs[0](x).unsqueeze(dim=-1)
for i in range(1, len(self.dilations)):
_y = self.convs[i](x).unsqueeze(dim=-1)
y = torch.cat((y, _y), dim=-1)
B, C, W, H, N = y.shape
if self.op == 'sum':
y = y.sum(dim=-1).flatten(2).permute(0,2,1).contiguous()
elif self.op == 'cat':
y = y.permute(0,4,1,2,3).flatten(3).reshape(B, N*C, W*H).permute(0,2,1).contiguous()
else:
raise NotImplementedError('no such operation: {} for multi-levels!'.format(self.op))
return y, (W, H)
class ReductionCell(nn.Module):
def __init__(self, img_size=224, in_chans=3, embed_dims=64, token_dims=64, downsample_ratios=4, kernel_size=7,
num_heads=1, dilations=[1,2,3,4], share_weights=False, op='cat', tokens_type='performer', group=1,
drop=0., attn_drop=0., drop_path=0., mlp_ratio=1.0):
super().__init__()
self.img_size = img_size
self.op = op
self.dilations = dilations
self.num_heads = num_heads
self.embed_dims = embed_dims
self.token_dims = token_dims
self.in_chans = in_chans
self.downsample_ratios = downsample_ratios
self.kernel_size = kernel_size
self.outSize = img_size
PCMStride = []
residual = downsample_ratios // 2
for _ in range(3):
PCMStride.append((residual > 0) + 1)
residual = residual // 2
assert residual == 0
self.pool = None
if tokens_type == 'pooling':
PCMStride = [1, 1, 1]
self.pool = nn.MaxPool2d(downsample_ratios, stride=downsample_ratios, padding=0)
tokens_type = 'transformer'
self.outSize = self.outSize // downsample_ratios
downsample_ratios = 1
self.PCM = nn.Sequential(
nn.Conv2d(in_chans, embed_dims, kernel_size=(3, 3), stride=PCMStride[0], padding=(1, 1), groups=group), # the 1st convolution
nn.SiLU(inplace=True),
nn.Conv2d(embed_dims, embed_dims, kernel_size=(3, 3), stride=PCMStride[1], padding=(1, 1), groups=group), # the 1st convolution
nn.BatchNorm2d(embed_dims),
nn.SiLU(inplace=True),
nn.Conv2d(embed_dims, token_dims, kernel_size=(3, 3), stride=PCMStride[2], padding=(1, 1), groups=group), # the 1st convolution
nn.SiLU(inplace=True))
self.PRM = PRM(img_size=img_size, kernel_size=kernel_size, downsample_ratio=downsample_ratios, dilations=self.dilations,
in_chans=in_chans, embed_dim=embed_dims, share_weights=share_weights, op=op)
self.outSize = self.outSize // downsample_ratios
in_chans = self.PRM.out_chans
if tokens_type == 'performer':
assert num_heads == 1
self.attn = Token_performer(dim=in_chans, in_dim=token_dims, head_cnt=num_heads, kernel_ratio=0.5)
elif tokens_type == 'performer_less':
self.attn = None
self.PCM = None
elif tokens_type == 'transformer':
self.attn = Token_transformer(dim=in_chans, in_dim=token_dims, num_heads=num_heads, mlp_ratio=mlp_ratio, drop=drop, attn_drop=attn_drop, drop_path=drop_path)
self.num_patches = (img_size // 2) * (img_size // 2) # there are 3 sfot split, stride are 4,2,2 seperately
def forward(self, x):
if len(x.shape) < 4:
B, N, C = x.shape
n = int(np.sqrt(N))
x = x.view(B, n, n, C).contiguous()
x = x.permute(0, 3, 1, 2)
if self.pool is not None:
x = self.pool(x)
PRM_x, _ = self.PRM(x)
if self.attn is None:
return PRM_x
convX = self.PCM(x)
x = self.attn.attn(self.attn.norm1(PRM_x))
convX = convX.permute(0, 2, 3, 1).view(*x.shape).contiguous()
x = x + convX
x = x + self.attn.drop_path(self.attn.mlp(self.attn.norm2(x)))
return x
class NormalCell(nn.Module):
def __init__(self, dim, num_heads, mlp_ratio=4., qkv_bias=False, qk_scale=None, drop=0., attn_drop=0.,
drop_path=0., act_layer=nn.GELU, norm_layer=nn.LayerNorm, class_token=False, group=64, tokens_type='transformer'):
super().__init__()
self.norm1 = norm_layer(dim)
self.class_token = class_token
if tokens_type == 'transformer':
self.attn = Attention(
dim, num_heads=num_heads, qkv_bias=qkv_bias, qk_scale=qk_scale, attn_drop=attn_drop, proj_drop=drop)
elif tokens_type == 'performer':
self.attn = AttentionPerformer(
dim, num_heads=num_heads, qkv_bias=qkv_bias, qk_scale=qk_scale, attn_drop=attn_drop, proj_drop=drop)
self.drop_path = DropPath(drop_path) if drop_path > 0. else nn.Identity()
self.norm2 = norm_layer(dim)
mlp_hidden_dim = int(dim * mlp_ratio)
self.mlp = Mlp(in_features=dim, hidden_features=mlp_hidden_dim, act_layer=act_layer, drop=drop)
self.PCM = nn.Sequential(
nn.Conv2d(dim, mlp_hidden_dim, 3, 1, 1, 1, group),
nn.BatchNorm2d(mlp_hidden_dim),
nn.SiLU(inplace=True),
nn.Conv2d(mlp_hidden_dim, dim, 3, 1, 1, 1, group),
nn.BatchNorm2d(dim),
nn.SiLU(inplace=True),
nn.Conv2d(dim, dim, 3, 1, 1, 1, group),
nn.SiLU(inplace=True),
)
def forward(self, x):
b, n, c = x.shape
if self.class_token:
n = n - 1
wh = int(math.sqrt(n))
convX = self.drop_path(self.PCM(x[:, 1:, :].view(b, wh, wh, c).permute(0, 3, 1, 2).contiguous()).permute(0, 2, 3, 1).contiguous().view(b, n, c))
x = x + self.drop_path(self.attn(self.norm1(x)))
x[:, 1:] = x[:, 1:] + convX
else:
wh = int(math.sqrt(n))
convX = self.drop_path(self.PCM(x.view(b, wh, wh, c).permute(0, 3, 1, 2).contiguous()).permute(0, 2, 3, 1).contiguous().view(b, n, c))
x = x + self.drop_path(self.attn(self.norm1(x)))
x = x + convX
x = x + self.drop_path(self.mlp(self.norm2(x)))
return x
Figure 4是grad-CAM,会有一些不同,但个人私货环节,可能并不是每个样本都会之前的方法好。

效果如下图table 2,引入了3个stage的CNN,将224*224做完token送进多头注意力机制的token数量降低了,因此参数量降低是很直观的,分类任务上效果还可以。

训练的配方:输入224*224, AdamW,用cosine learning rate scheduler,初始学习率0.0001,batch size是512, 数据增强方法跟T2T一样即mixup和cutmix。针对ViT难以训练的毛病,表现了ViTAE能够在小的数据集中收敛和训练速度,这部分在个人觉得在加入了CNN后,就会有这方面的增益。
平均注意力距离:Figure 3做了一个挺有意思的分析,average attention distance of each layer,每层平均注意力距离。ViTAE在浅层(1-3层,也就是RCs)的长距离依赖关系弱于深层(即4-7层,也就是NCs)。说明了训练完的模型,确实跟一开始的设定一样。但我不是很同意文中的表述(It can be observed that with the usage of PCM, which focuses on modeling locality, the transformer layers in the proposed NCs can better focus on modeling long-range dependencies, especially in shallow layers.),因为NC也有PCM模块。

个人思考:ConvNext将Swin Transformer一步步用CNN来替换,也获得非常不错的效果。实际上使用的时候,个人认为并不用关心它是transformer还是CNN,只要相应的算子能支持,在算力、功耗、指标等限制下,能跑得更快更好那就可以了。ConvNext侧面证明了一点,纯transformer的架构并没有遥遥领先与CNN架构,那么如何将CNN和transformer有机的结合在一起,在更具体的任务中能够提点,是我关心的要点。ViTAE也是CNN和tranformer结合的一种尝试,与其说是CNN结合transformer,我更愿意理解为:拿更有效的token送进self-attention中去建模
